
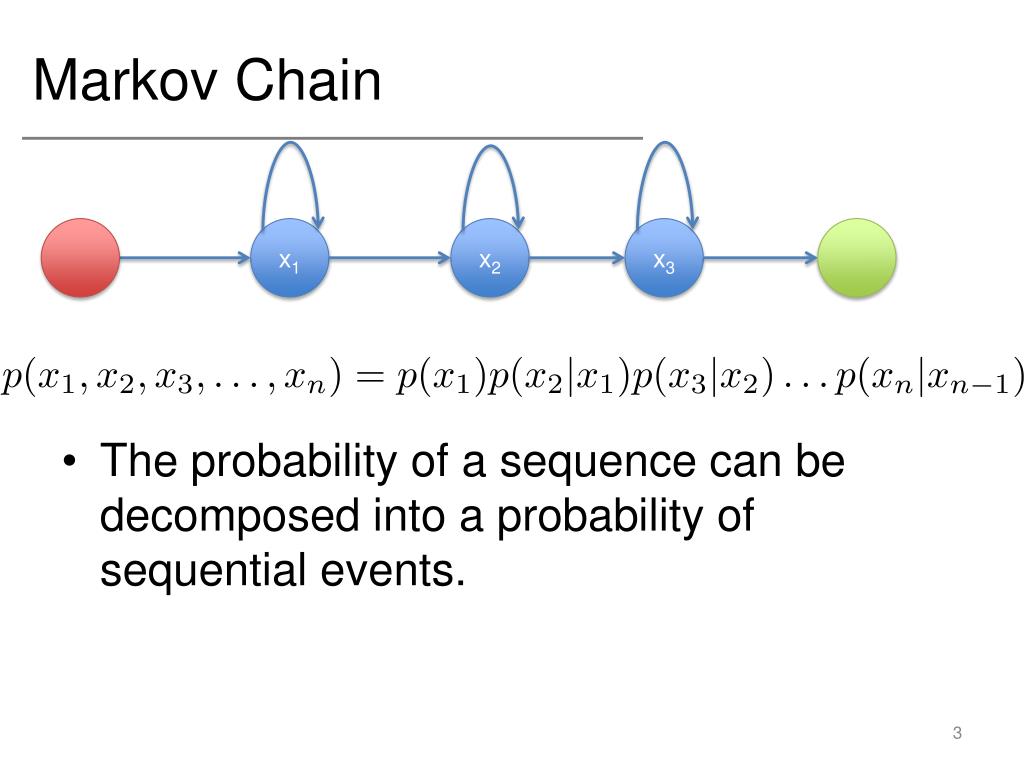
Note that the COUNT is designated as a weight variable for the GENLOG analyses but this designation is turned off for the CNLR comand. The Value of Loss Function reports the G^2 result and the parameter estimates are reported in the various Parameter columns.ĬNLR was used for the last model because of its capability to constrain parameters (to be equal in this case). In the CNLR output, look at the last row of the Iteration History table (Iteration 18.1). You can find the goodness of fit results (to compare to Agresti's G^2) in the Likelihood Ratio row of the Goodness-of-Fit table of the GENLOG output. The goodness of fit results match those from Agresti's text and the parameter estimates for the last 2 models match those in Agresti's Table 11.8.Īgresti does not report the parameter estimates for the first 2 models, the first-order and second-order Markov chains, which both fit poorly. The GENLOG and CNLR commands below run all four models reported on pages 478-479 of Agresti. GENLOG would have produced the same results with the (1,2) coding in Agresti's data table. The data differs from Agresti's table only in that variables X9 to X12 are coded as 0 and 1, rather than as 1 and 2. The variable COUNT holds the number of subjects observed at each sequence. The data are reported in an aggregated form, where each case represents one of the 16 possible sequences of values for the four variables. In this example, variables X9, X10,X11,and X12 are binary variables that represent the presence (1) or absence (0) of respiratory illness in children that were observed each year at ages 9 to 12. The following SPSS commands illustrate the use of the SPSS procedures GENLOG and CNLR (Constrained NonLinear Regression) to perform first- and second-order Markov chains and a pair of alternate models.


 0 kommentar(er)
0 kommentar(er)
